38 Lecture 502
Regression
38.1 Regression analysis
Regression analysis is a supervised machine learning approach
Predict the value of one outcome variable as
\[outcome_i = (model) + error_i \]
- one predictor variable (simple / univariate regression)
\[Y_i = (b_0 + b_1 * X_{i1}) + \epsilon_i \]
- more predictor variables (multiple / multivariate regression)
\[Y_i = (b_0 + b_1 * X_{i1} + b_2 * X_{i2} + \dots + b_M * X_{iM}) + \epsilon_i \]
38.2 Least squares
Least squares is the most commonly used approach to generate a regression model
The model fits a line
- to minimise the squared values of the residuals (errors)
- that is squared difference between
- observed values
- model
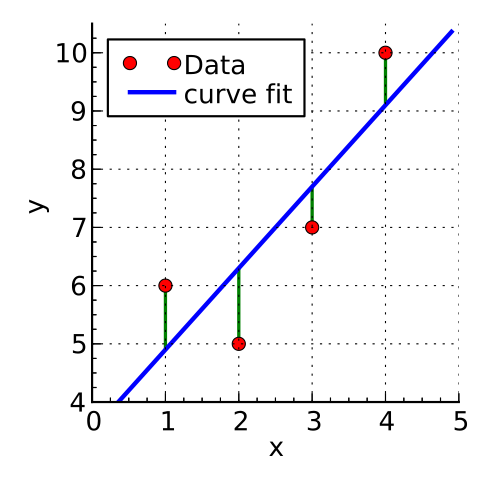
by Krishnavedala
via Wikimedia Commons,
CC-BY-SA-3.0
\[deviation = \sum(observed - model)^2\]
38.3 Example
\[arr\_delay_i = (b_0 + b_1 * dep\_delay_{i1}) + \epsilon_i \]
##
## Call:
## lm(formula = arr_delay ~ dep_delay)
##
## Residuals:
## Min 1Q Median 3Q Max
## -43.906 -9.022 -1.758 8.678 57.052
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -4.96717 0.43748 -11.35 <2e-16 ***
## dep_delay 1.04229 0.01788 58.28 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 13.62 on 972 degrees of freedom
## Multiple R-squared: 0.7775, Adjusted R-squared: 0.7773
## F-statistic: 3397 on 1 and 972 DF, p-value: < 2.2e-1638.4 Overall fit
The output indicates
- p-value: < 2.2e-16: \(p<.001\) the model is significant
- derived by comparing the calulated F-statistic value to F distribution 3396.74 having specified degrees of freedom (1, 972)
- Report as: F(1, 972) = 3396.74
- Adjusted R-squared: 0.7773: the departure delay can account for 77.73% of the arrival delay
- Coefficients
- Intercept estimate -4.9672 is significant
dep_delay(slope) estimate 1.0423 is significant
38.5 Parameters
\[arr\_delay_i = (Intercept + Coefficient_{dep\_delay} * dep\_delay_{i1}) + \epsilon_i \]
flights_nov_20 %>%
ggplot(aes(x = dep_delay, y = arr_delay)) +
geom_point() + coord_fixed(ratio = 1) +
geom_abline(intercept = 4.0943, slope = 1.04229, color="red")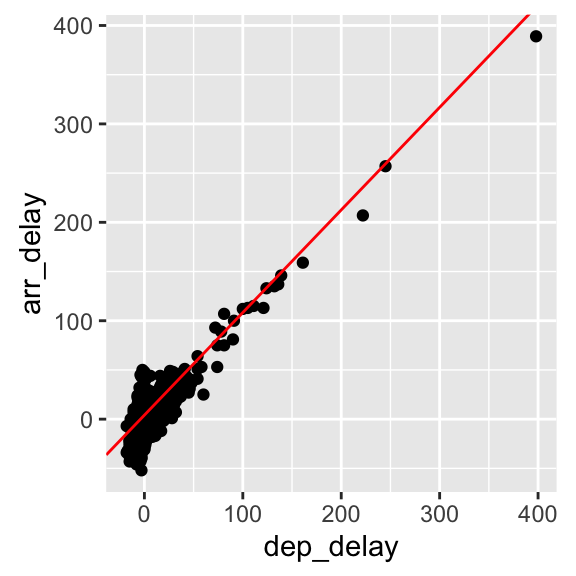
38.6 Checking assumptions
- Linearity
- the relationship is actually linear
- Normality of residuals
- standard residuals are normally distributed with mean
0
- standard residuals are normally distributed with mean
- Homoscedasticity of residuals
- at each level of the predictor variable(s) the variance of the standard residuals should be the same (homo-scedasticity) rather than different (hetero-scedasticity)
- Independence of residuals
- adjacent standard residuals are not correlated
- When more than one predictor: no multicollinearity
- if two or more predictor variables are used in the model, each pair of variables not correlated
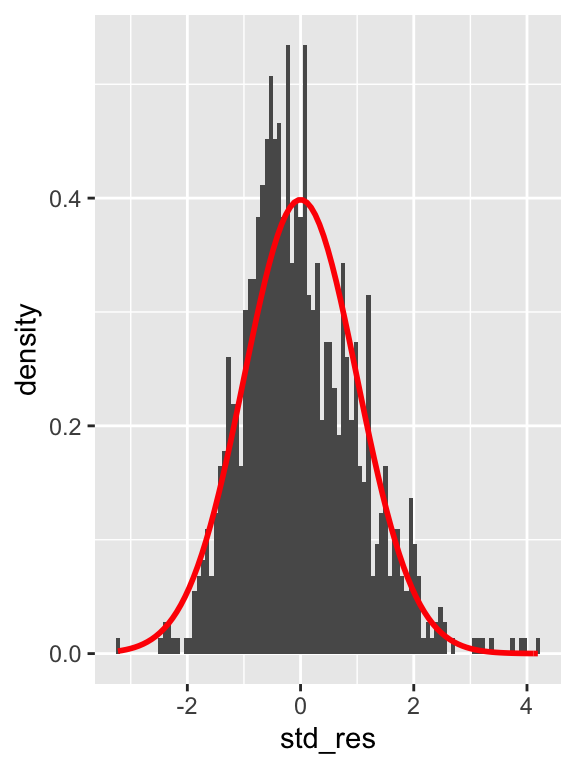
38.7 Normality
Shapiro-Wilk test for normality of standard residuals,
- robust models: should be not significant
##
## Shapiro-Wilk normality test
##
## data: .
## W = 0.98231, p-value = 1.73e-09Standard residuals are NOT normally distributed
38.8 Homoscedasticity
Breusch-Pagan test for homoscedasticity of standard residuals
- robust models: should be not significant
##
## studentized Breusch-Pagan test
##
## data: .
## BP = 0.017316, df = 1, p-value = 0.8953Standard residuals are homoscedastic
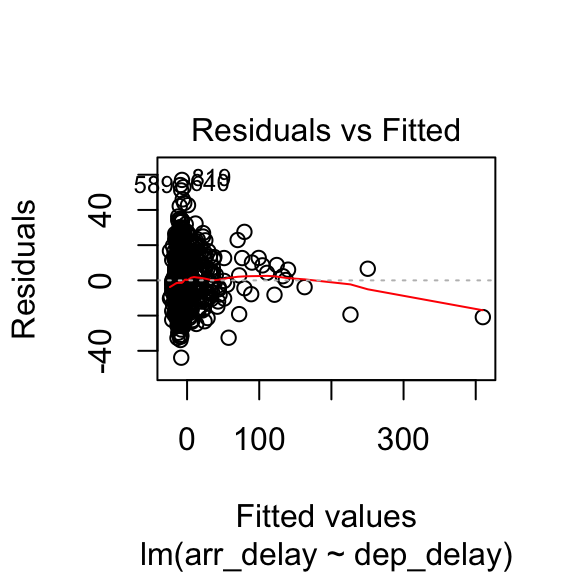
38.9 Independence
Durbin-Watson test for the independence of residuals
- robust models: statistic should be close to 2 (between 1 and 3) and not significant
##
## Durbin-Watson test
##
## data: .
## DW = 1.8731, p-value = 0.02358
## alternative hypothesis: true autocorrelation is greater than 0Standard residuals might not be completely indipendent
Note: the result depends on the order of the data.